By the Office of Engineering Communications
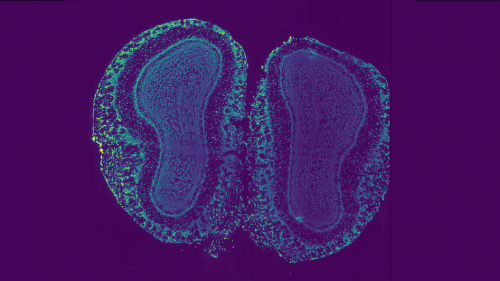
Using a new deep learning algorithm, researchers at Princeton have designed an approach to map the cellular organization of tissues by modeling spatial patterns in molecular data. This development has implications for advancing cancer treatment and understanding aging, development and other cellular processes.
“Biological tissues are organized into intricate spatial structures that are important for their function,” said Ben Raphael, Class of 1991 Graduate Professor of Computer Science. “But it’s very hard to directly measure tissue organization from molecular data. This method reveals the hidden geometry.”
Every part of our bodies is made up of cells, and the function of each cell is determined by thousands of genes that are active within it. While powerful genetic sequencing tools can give researchers information on exactly which genes are being expressed in a piece of tissue, these older technologies could not measure the locations of cells.

"This sequencing data is like having thousands of mixed-up puzzle pieces,” said Raphael. “Each piece is full of information, but you don’t know how it connects to the other pieces, and many of the pieces are missing.” Sequencing a tumor, for example, might reveal which cancer cells are present, but it won’t show exactly where in the tumor these cancer cells are located.
In a paper published earlier this year in Nature Methods, Raphael and his co-authors showed how to use a newer technology called spatial transcriptomics to take these jumbled pieces — measurements of mRNA across thousands of genes — and assemble them into a topographic map of gene expression across a piece of tissue.
The algorithm they created to do this uses deep learning, a kind of artificial intelligence, that reveals subtle variation in gene expression across tissues.
According to Raphael, the topographic map is particularly useful for research on tumors, which are a mix of healthy and abnormal cells. Understanding how tumors work on a cellular level can reveal where cancer cells are growing and whether certain non-cancerous cells are contributing to tumor growth. “There are all kinds of mysteries related to the tumor microenvironment that could be revealed,” said Raphael.
The research also reveals fundamental insights into how cells behave, said Uthsav Chitra, the first author on the paper, who is a 2024 Princeton graduate alumnus and now an assistant professor at Johns Hopkins University. Because the map reveals a gradient of gene expression across cells, it can show how cells change, interact, and migrate as they mature. Understanding how cells vary in a tissue is the first step to controlling cell behavior, Chitra said.
While the algorithm was initially developed for spatial transcriptomics data, Raphael and his group members knew that the approach was adaptable to other types of data. They didn’t have a long wait to test this idea: in a collaboration with Joshua Rabinowitz, professor of chemistry and the Lewis-Sigler Institute for Integrative Genomics and director of the Princeton Branch of the Ludwig Institute for Cancer Research, they adapted the algorithm to analyze spatial patterns of metabolites – small molecules such as sugars – that are important for cellular function.
In a paper published Oct. 15 in Nature they mapped changes in metabolites within the liver and small intestine, a first step toward examining how metabolism changes in aging and disease.
"We are excited to push further applications of this approach with new spatial profiling technologies,” said Raphael.
The article, Mapping the topography of spatial gene expression with interpretable deep learning, was published in Nature Methods on January 23, 2025. Besides Raphael and Chitra, authors include Brian Arnold, Hirak Sarkar, Kohei Sanno, and Sereno Lopez-Darwin from Princeton University and Cong Ma from the University of Michigan Medical School. Support was provided by the Ludwig Institute for Cancer Research, Schmidt Futures Foundation, National Cancer Institute (NCI), National Science Foundation (NSF) and the Damon Runyon Cancer Research Foundation.
The article, Spatial metabolic gradients in the liver and small intestine, was published in Nature on October 15, 2025. In addition to Raphael, Chitra and Rabinowitz, authors include Laith Z. Samarah, Clover Zheng, Xi Xing, Wong Dong Lee, Amichay Afriat, Michael MacArthur, Wenyun Lu, Connor S.R. Jankowski, Cong Ma, Craig J. Hunter, Michael Neinast, and Daniel R. Weilandt of Princeton University.