Ellen D. Zhong

Email: zhonge [at] princeton.edu
Office: 314 Computer Science
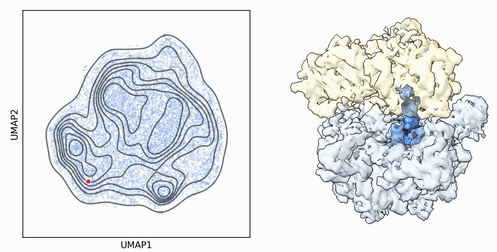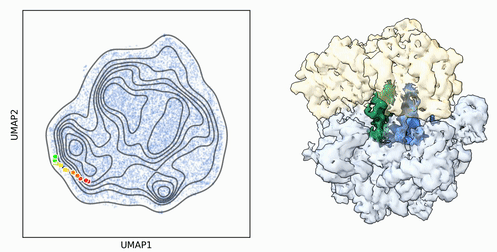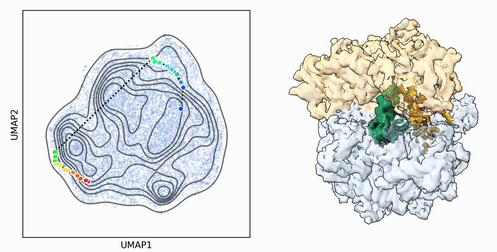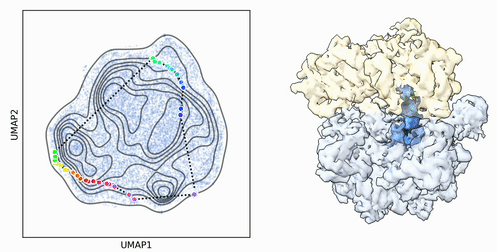
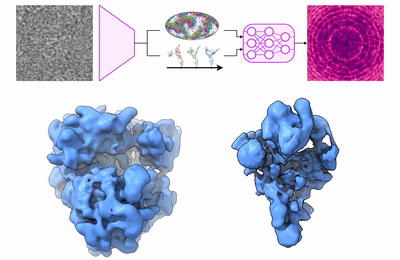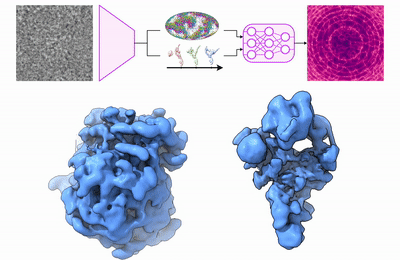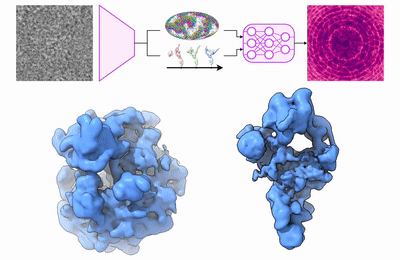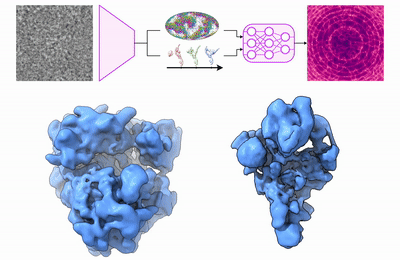
I am an Assistant Professor of Computer Science at Princeton University where I lead the E.Z. Lab for Molecular Machine Learning. We are interested in problems at the intersection of AI and structural biology. In particular, our group focuses on developing methods for image analysis and 3D reconstruction that enable new discoveries in protein structure, dynamics, and interactions. For more information about our research, please visit our lab website.
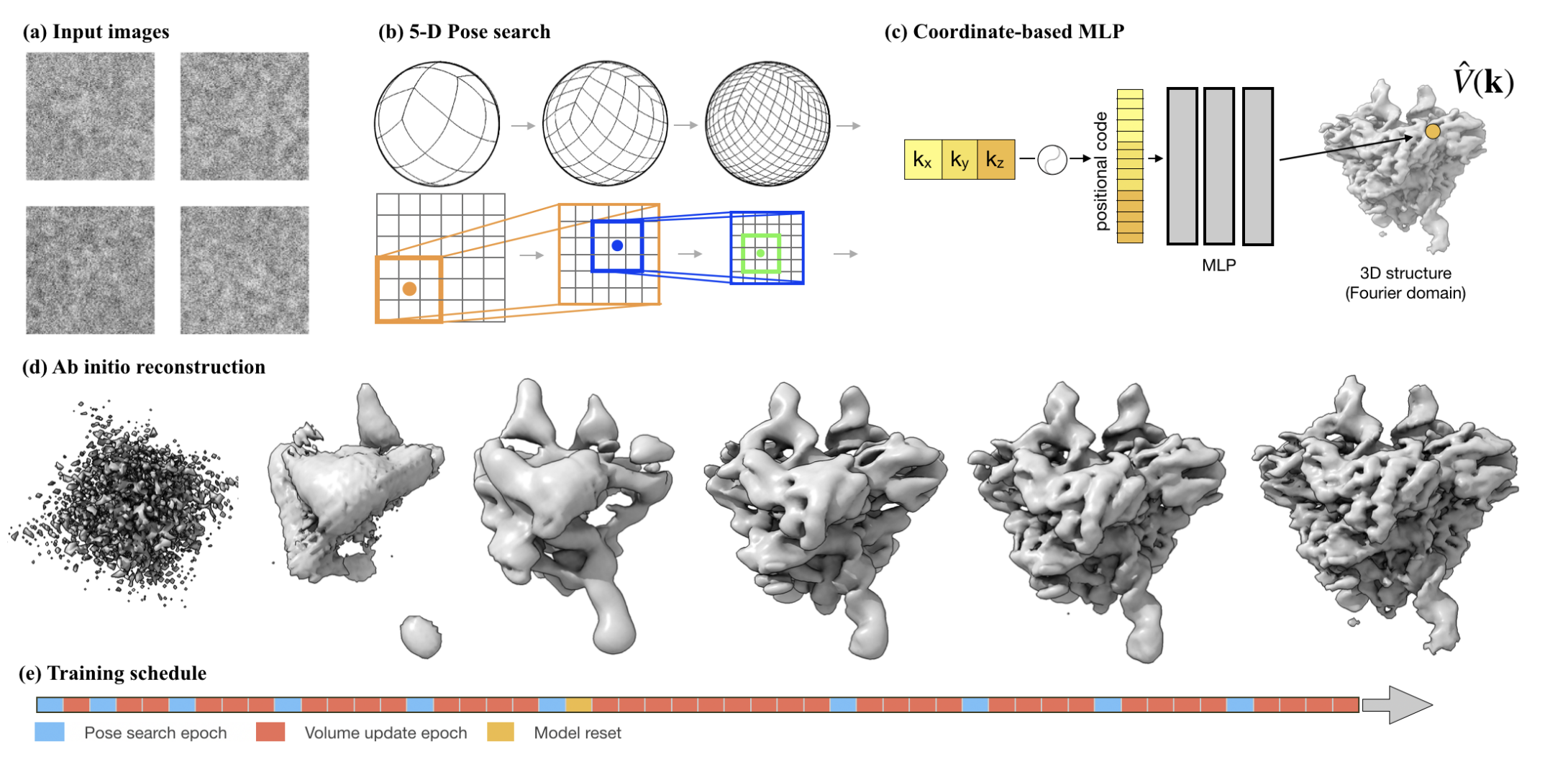
Previously, I obtained my Ph.D. at MIT in 2022, where I developed deep learning methods for 3D reconstruction of dynamic protein structures from cryo-EM images. I have interned at DeepMind on the AlphaFold team, and prior to my Ph.D., I worked at D. E. Shaw Research on molecular dynamics algorithms for drug discovery.