Ellen D. Zhong

Email: zhonge [at] princeton.edu
Office: 314 Computer Science
I am an Assistant Professor of Computer Science at Princeton University, where I am also affiliated with the Princeton Laboratory for Artificial Intelligence, the Center for Statistics and Machine Learning, the Omenn-Darling Bioengineering Institute, and the Department of Molecular Biology. I am in interested in problems at the intersection of AI and the molecular sciences with the goal of developing methods that enable new scientific discoveries.
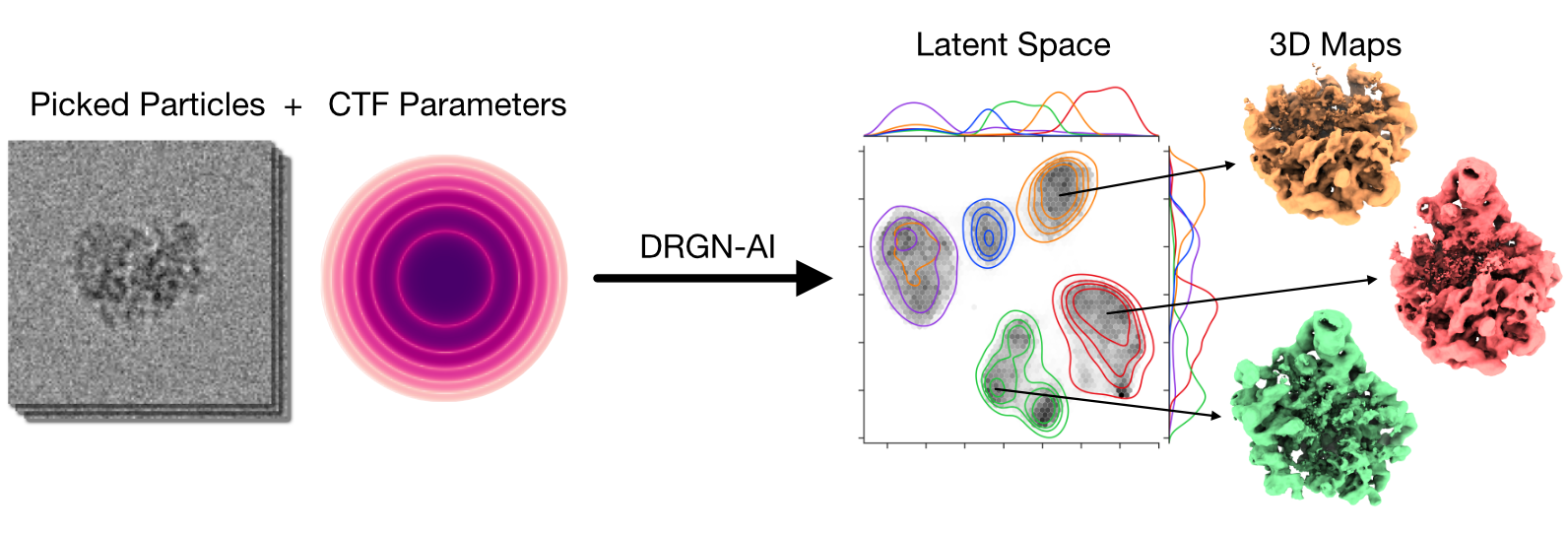
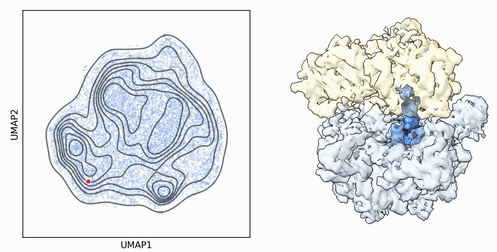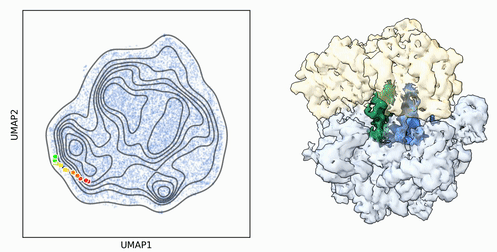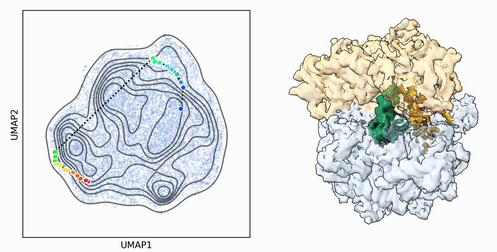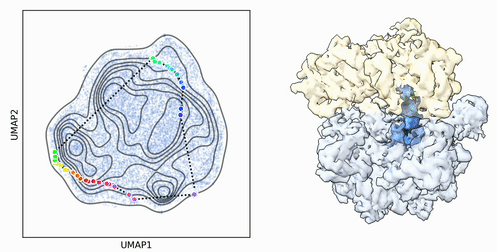
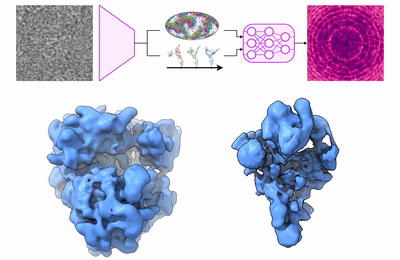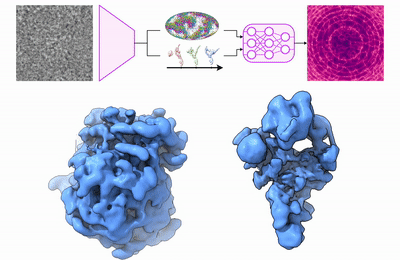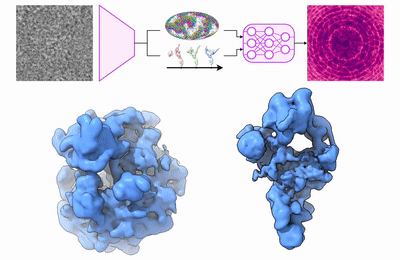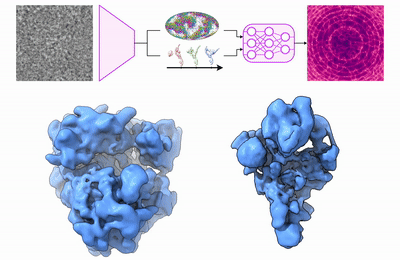
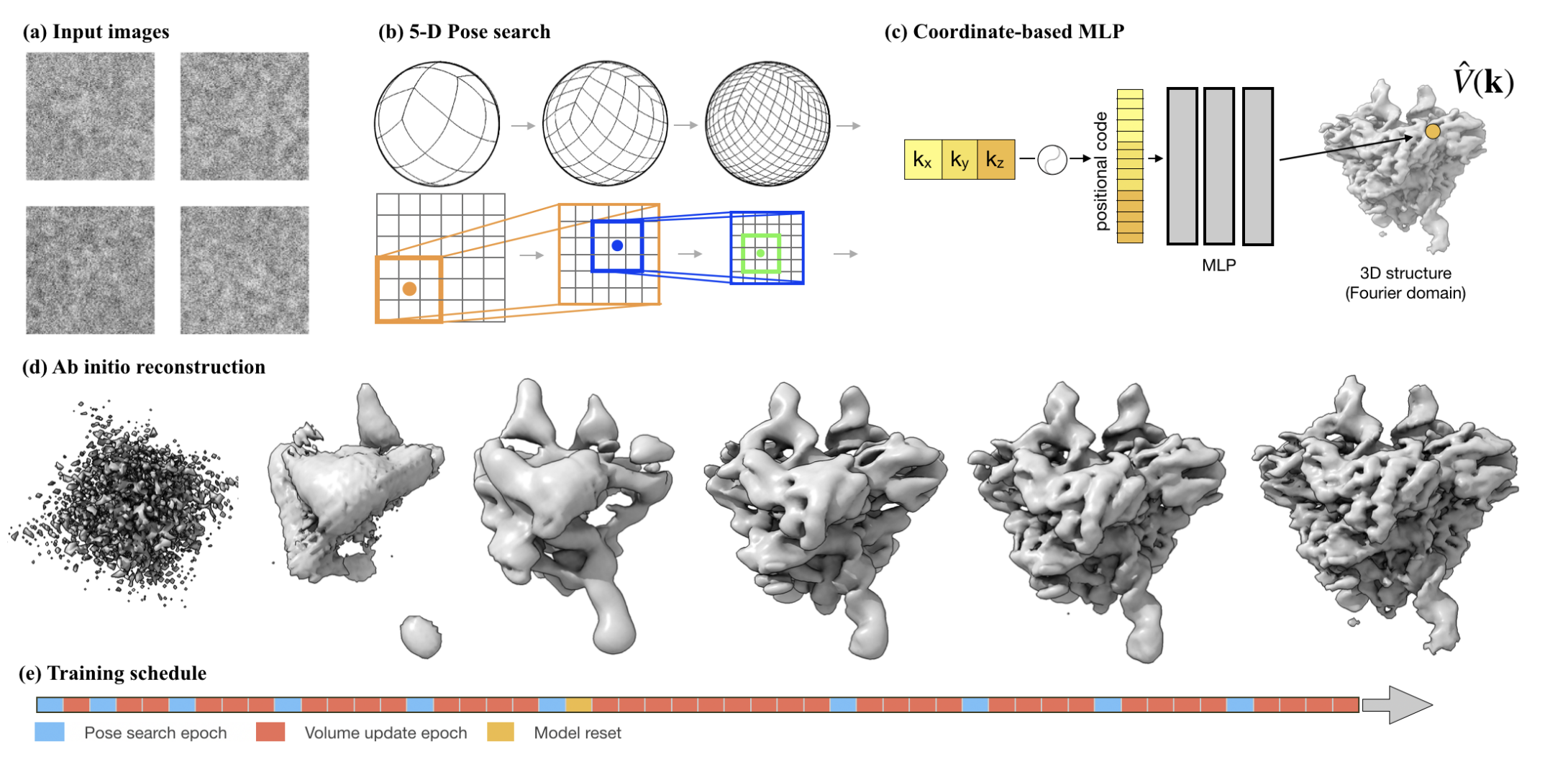
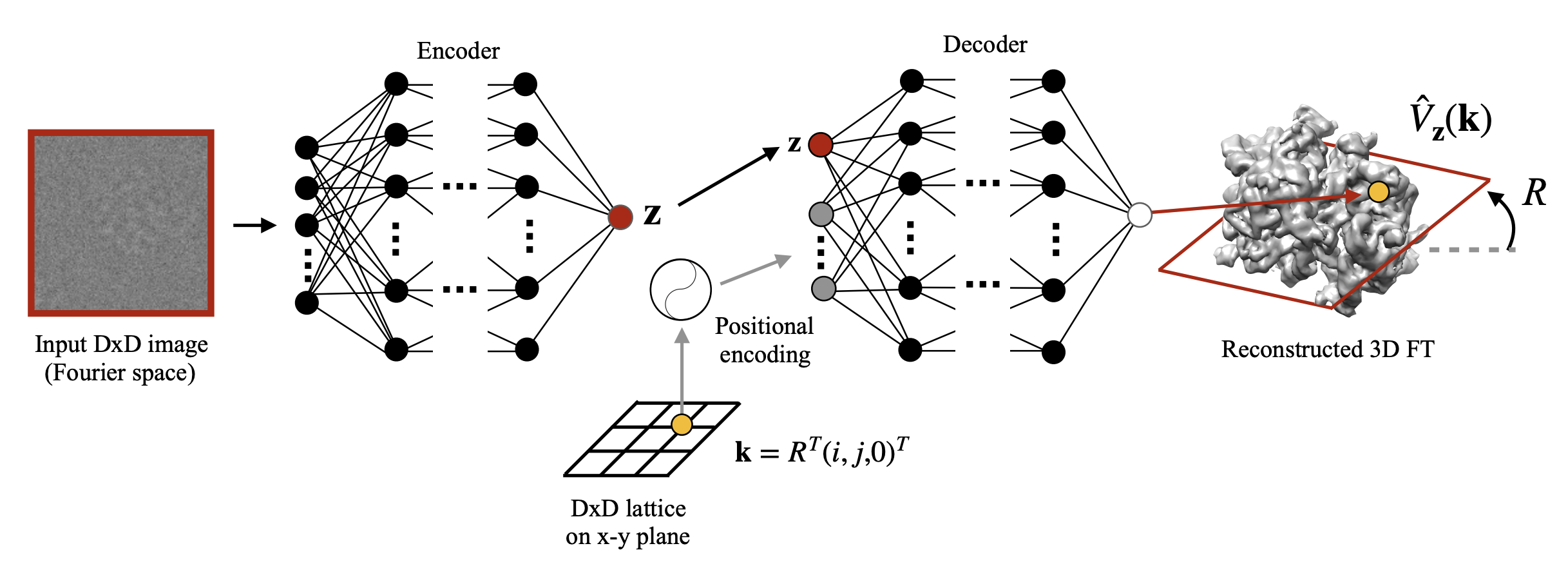
My research program spans methodological research in AI and computer vision, as well as close collaboration with experimentalists in molecular biology and chemistry. A focus of my group is developing deep learning methods for 3D reconstruction of protein structure from imaging data, and we develop open-source software in active use by the structural biology community.
Our research has been recognized with the NIH Director’s New Innovator Award, the Schmidt Sciences AI2050 Early Career Fellowship, a Major Society Award from the Microscopy Society of America, the Eric and Wendy Schmidt Transformative Technology Fund Award, and the E. Lawrence Keyes, Jr./Emerson Electric Co. Faculty Advancement Award from Princeton University. I am grateful for support from the NIH, the Chan Zuckerberg Initiative, Schmidt Sciences, Janssen Pharmaceuticals, Generate Biomedicines, and Princeton University.
I received my Ph.D. from MIT in 2022 before joining the Princeton faculty. Previously, I have worked at Google DeepMind on the AlphaFold team, and at D. E. Shaw Research on molecular dynamics algorithms for drug discovery.
I also spend some time with companies and research institutes. My current industry and professional engagements include:
- Generate Biomedicine (Professor in Residence)
- The Chan Zuckerberg Imaging Institute (Scientific Advisory Board)
- Deep Apple Therapeutics (Scientific Advisory Board)
CV / Scholar / GitHub / Twitter / Lab
Now recruiting graduate students and postdocs for Fall 2026! Our group aims to define new problems for AI in molecular biology and chemistry. We work at the intersection of many areas including 3D vision & generative modeling for proteins. Please see our lab website for more information.