| repeats | diagnosis |
| 0–9 | not human |
| 10–35 | normal |
| 36–39 | high risk |
| 40–180 | Huntington's disease |
| 181–∞ | not human |
Write a data type Huntingtons that takes a human DNA sequence and determines whether the patient is at risk for HD by determining the maximum number of CAG repeats. Implement the following API:
public class Huntingtons
---------------------------------------------------------------------------
Huntingtons(String dna) // construct a new object using given DNA
int maxRepeats() // return the maximum number of CAG repeats
String diagnosis() // return the diagnosis as a String
- Constructor. The constructor takes a human DNA sequence as an argument. It should throw a RuntimeException if any character in the string is not an (uppercase) 'A', 'C', 'T', 'G', or 'N' or if there are two or more consecutive 'N' characters. The characters 'A', 'C', 'T', and 'G' represent the nucleic acids Adenine, Thymine, Guanine, and Cytosine, respectively. The character 'N' indicates that the nucleic acid is unknown.
- Maximum number of CAG repeats. The maxRepeats() method returns the maximum number of CAG repeats in the DNA sequence. Consider 'N' to be a wildcard character; for example, CAGCANCAGNAG represents a CAG repeat of size 4.
- Medical diagnosis. The diagnosis() method returns the medical diagnosis as a string, using the exact text in the table above.
For full credit, your program must use regular expressions to find the CAG repeats. Also, include a test client main() that takes a sequence of filenames as command-line arguments, and outputs the maximum number of CAG repeats and the corresponding medical diagnosis for each.
The data file format will consist of a sequence of the characters 'A', 'C', 'T', 'G', or 'N', along with arbitrary amounts of intervening whitespace (which you will need to strip out before invoking the constructor). For example, the DNA sequences in test4.txt, test5.txt, test64.txt, chromosome4-hd.txt, and chromosome4-healthy.txt have CAG repeats of sizes 4, 5, 64, 79, and 19, respectively.
% more test4.txt % more test64.txt
TTTTTTTTTT TTTTTTTTTT TTTTTTTTCA TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT
GCAGCAGCAG TTTCAGCAGT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTCAGCAGCA
TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
TTTCAGTTTT TTTTTTTTTT T GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
% more test5.txt GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
TTTTTTTTTT TTTTTTTTTT TTTTTTTTNA GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCNGNAGCAN CAGTTTCAGC AGCAGTTTTT GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT GCAGTTTTTT TTTTTTTTTT TTTTTTTTTT
TTTCAGTTTT TTTTTTTTTT T TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT
% java Huntingtons test4.txt test5.txt test64.txt chromosome4-hd.txt chromosome4-healthy.txt
test4.txt
max number of CAG repeats = 4
not human
test5.txt
max number of CAG repeats = 5
not human
test64.txt
max number of CAG repeats = 64
Huntington's disease
chromosome4-hd.txt
max number of CAG repeats = 79
Huntington's disease
chromosome4-healthy.txt
max number of CAG repeats = 19
normal
As an example, The Turing machine binaryadder.tur takes as input two binary integers (separated by the + symbol) and terminates with the sum written in the tape.
% more binaryadder.tur title Binary Adder description Kevin Wayne, wayne, P01 Add two binary numbers, and leave the result on the tape. vertices 2 R 0.25 0.75 0 L 0.25 0.25 1 L 0.75 0.25 3 L 0.75 0.75 4 R 0.40 0.50 5 H 0.60 0.50 edges 0 0 0 1 135 0 1 1 0 0 4 + # 1 3 + + 2 0 # # 3 2 # 1 0.2 3 2 0 1 -0.2 3 3 1 0 -30 4 4 1 # -135 4 5 # # tapes [1] 0 1 0 + 1 1 1 1 [1] 0 0 + 1 1 1 [1] 1 0 0 + 1 1 [1] 0 1 0 + 1 0 0 1 0 1 |
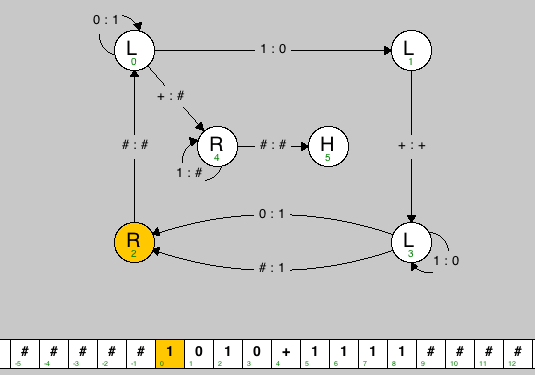 |
The data file is divided into 5 sections (title, description, vertices, edges, tapes), and each section begins with the corresponding name. Each vertex (state) is specified with 4 required values: its name (a string), its type (L, R, Y, N, H), and its x- and y-coordinates (between 0 and 1). The first state listed is the start state. Each edge (transition) is specified with 4 required values: the starting state, the ending state, the symbol to read, and the symbol to write. An optional 5th parameter specifies the curvature with which to draw the edge (straight line by default). Each sample input tape is specified by listing its symbols, separated by spaces. The tape head starts at the symbol delimited by square braces - for this assignment, it should always be the leftmost non-blank character.
Submission. Submit Huntingtons.java and comparator.tur. Also, submit a readme.txt and answer any questions.
Extra credit. If you based your comparator on binaryadder.tur, then it is very similar to a subtractor. Unfortunately, it would take more than 2n steps to subtract two n-bit numbers. Your challenge is to design a Turing machine that subtracts two positive n-bit numbers in time proportional to n2. Assume the left number on the tape is always larger than or equal to the right number, so the difference will always be greater than or equal to zero. However, do not assume that the two numbers always have the same number of bits. Name your file efficientSubtractor.tur and submit it as an Additional File.
This assignment was created by Kevin Wayne. Copyright © 2004.