| repeats | diagnosis |
| 0–9 | not human |
| 10–35 | normal |
| 36–39 | high risk |
| 40–180 | Huntington's disease |
| 181–∞ | not human |
Write a program Huntingtons.java to read in a human DNA sequence from a file and identify whether the patient is at risk for HD by determining the maximum number of CAG repeats. Output the maximum number of repeats, and an appropriate medical diagnosis based on the table above. The data file format will consist of a sequence of the letters A, C, G, and T, along with arbitrary amounts of whitespace separating the letters. For full credit, your program should use regular expressions and should identify CAG repeats, even if there is intervening whitespace.
For example, the DNA sequences in test4.txt, test64.txt, chromosome4-hd.txt, and chromosome4-healthy.txt have CAG repeats of sizes 4, 64, 79, and 19, respectively.
% more test4.txt % more test64.txt
TTTTTTTTTT TTTTTTTTTT TTTTTTTTCA TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT
GCAGCAGCAG TTTCAGCAGT TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT TTCAGCAGCA
TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
TTTCAGTTTT TTTTTTTTTT T GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGCAGCAG CAGCAGCAGC AGCAGCAGCA
GCAGTTTTTT TTTTTTTTTT TTTTTTTTTT
TTTTTTTTTT TTTTTTTTTT TTTTTTTTTT
% java Huntingtons test4.txt %java Huntingtons test64.txt
max number of CAG repeats = 4 max number of CAG repeats = 64
not human Huntington's disease
% java Huntingtons chromosome4-hd.txt % java Huntingtons chromosome4-healthy.txt
max number of CAG repeats = 79 max number of CAG repeats = 19
Huntington's disease normal
Download turing.jar and launch it by typing java -jar turing.jar from the command line in the command prompt window (windows) or terminal window (mac). Your job is to create and submit a text file parenbrack.tur containing the description of your Turing machine like paren.tur, but instead of matching just parentheses, you are to match both parentheses and angle brackets.
% more paren.tur title Parenthesis Matcher description Accepts the input if it is a well-balanced sequence of parentheses. vertices 0 R 0.2 0.5 1 L 0.5 0.5 2 L 0.5 0.9 3 Y 0.8 0.9 4 N 0.8 0.5 edges 0 1 ) X 0.2 1 0 ( X 0.2 0 2 # # 2 3 # # 2 4 ( ( 1 4 # # tapes [(] ) [(] [)] ( [(] ( ) ) ( ( ( ) ) ) ( ) [(] ( ( ( ) ) [(] ( ( ) ) ) [(] ) ) ( ( ) ( ) [(] ( ) ) ) ( ) ) |
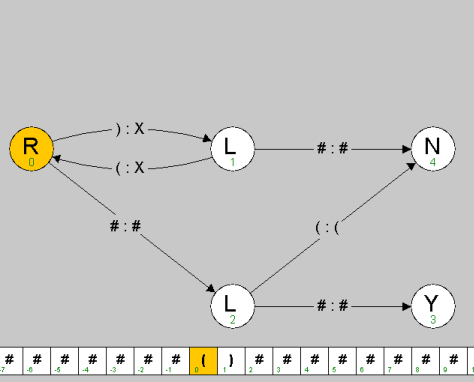 |
The data file is divided into 5 sections (title, description, vertices, edges, tapes), and each section begins with the corresponding name. Each vertex (state) is specified with 4 required values: its name (a string), its type (L, R, Y, N, H), and its x- and y-coordinates (between 0 and 1, where (0, 0) is the upper left corner). The first state listed is the start state. Each edge (transition) is specified with 4 required values: the starting state, the ending state, the symbol to read, and the symbol to write. An optional 5th parameter specifies the curvature with which to draw the edge (straight line by default). Each sample input tape is specified by listing its symbols, separated by spaces. The tape head starts at the symbol delimited by square braces - for this assignment, it should always be the leftmost non-blank character.
Submission. Submit Huntingtons.java and parenbrack.tur. Also, submit a readme.txt file and answer the questions.
This assignment was created by Kevin Wayne and Maia Ginsburg. Copyright © 2004.